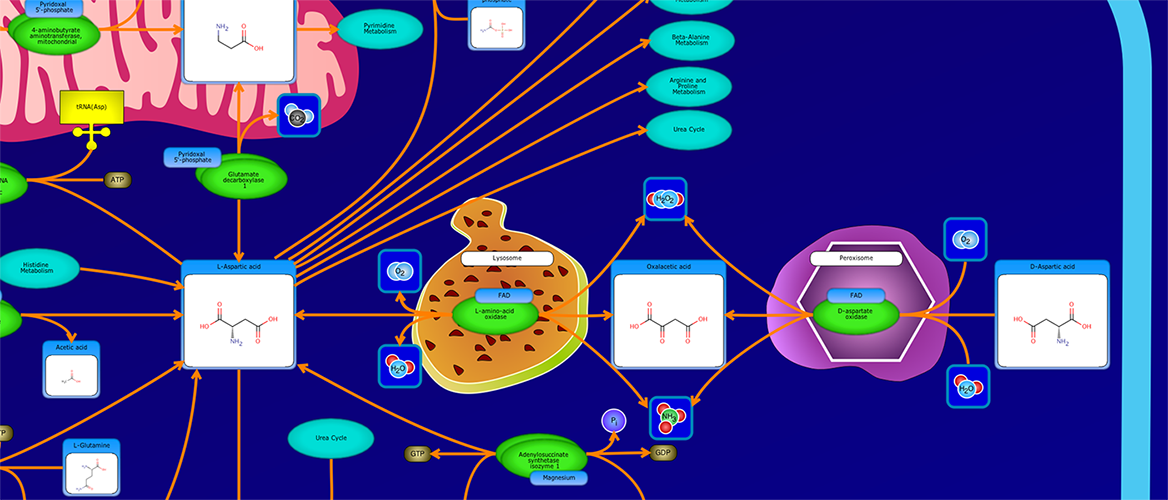
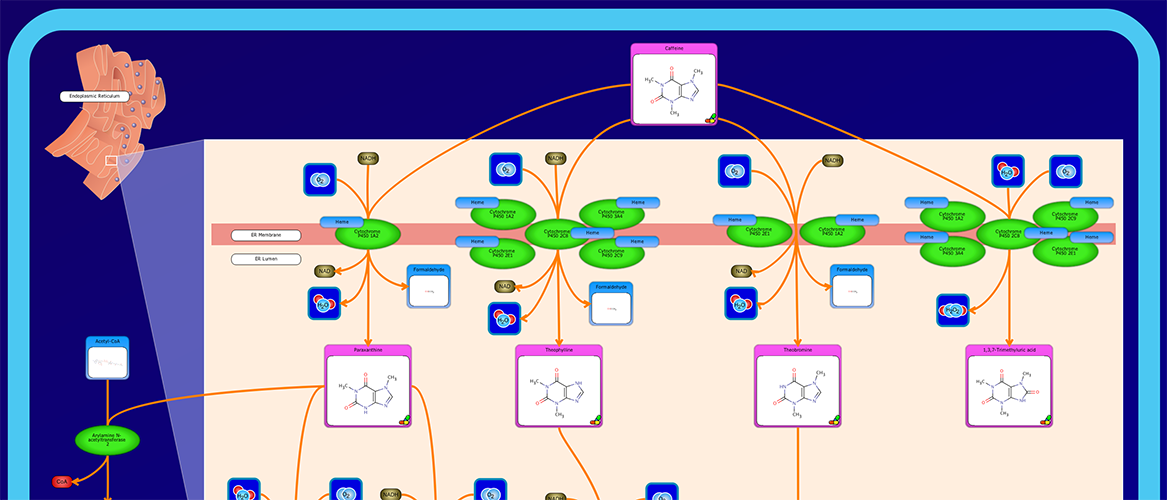
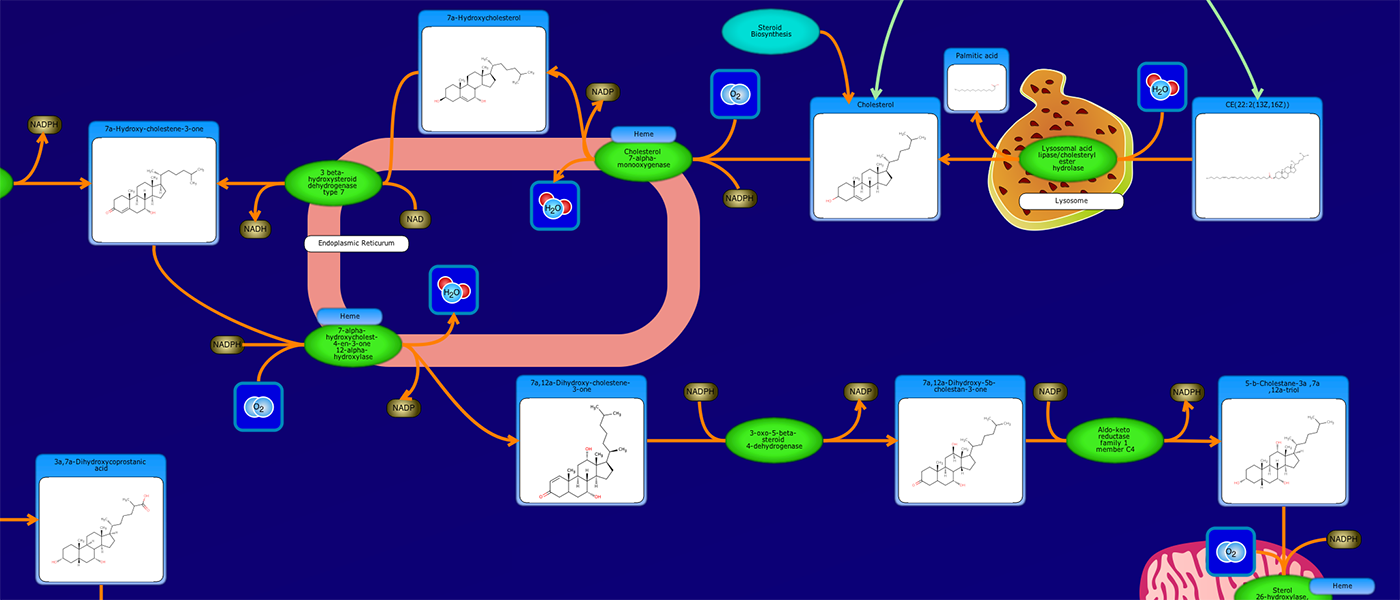
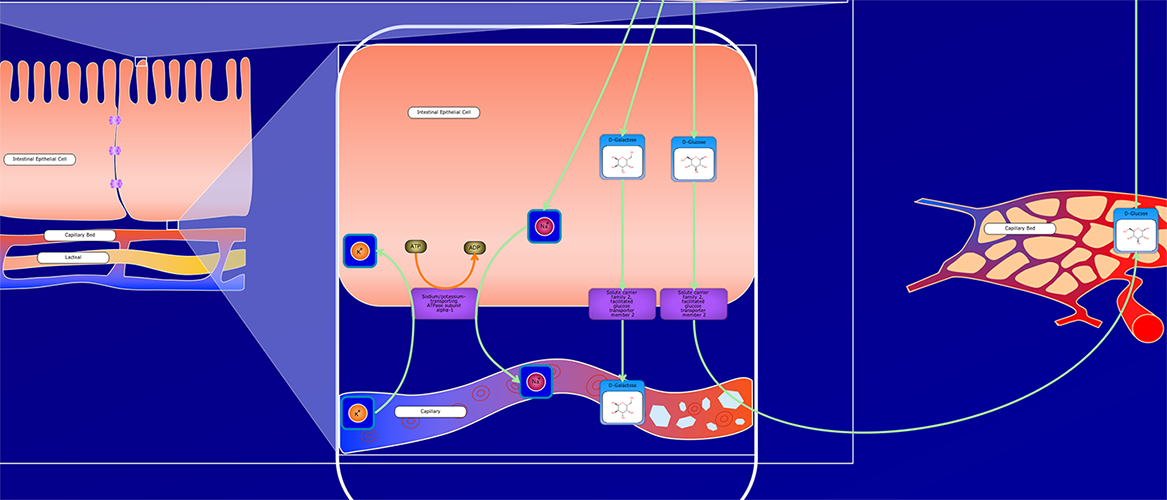
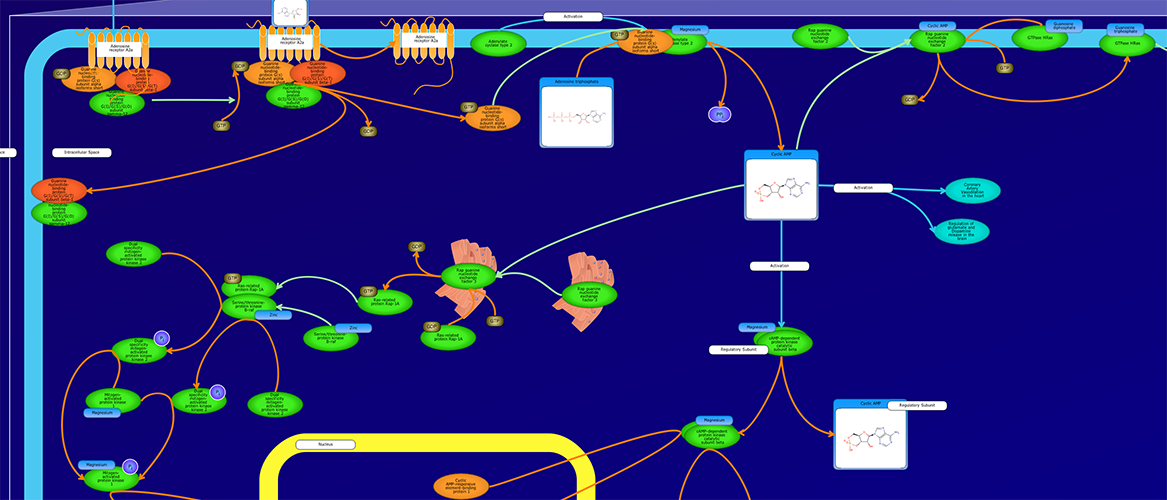
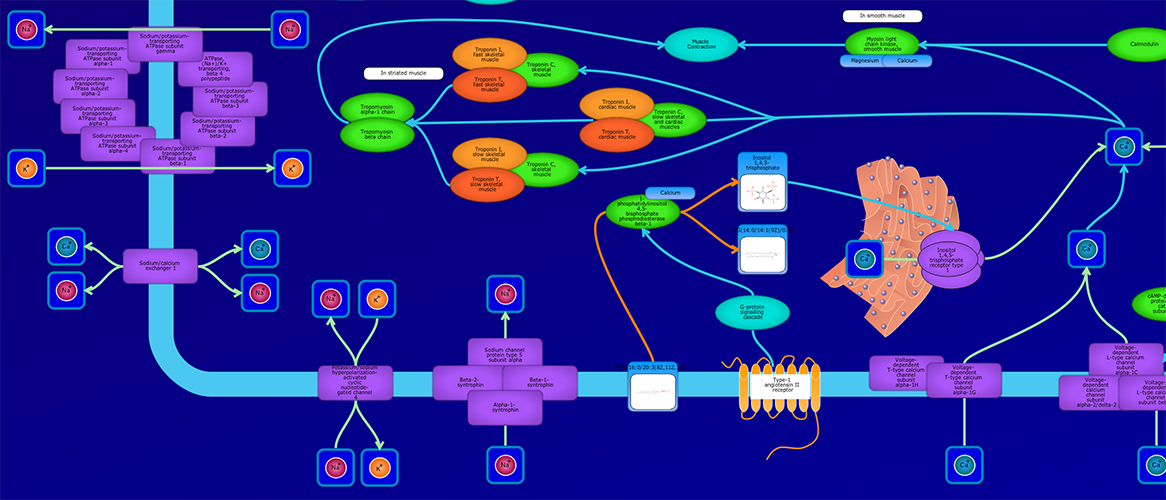
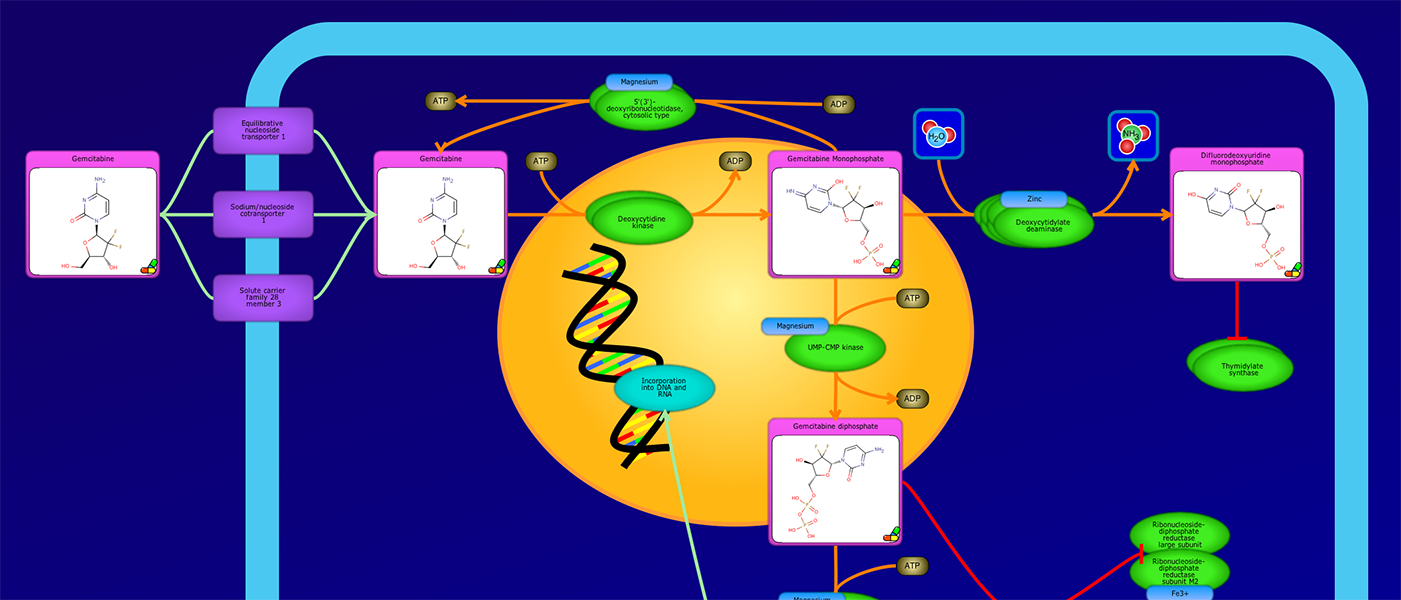
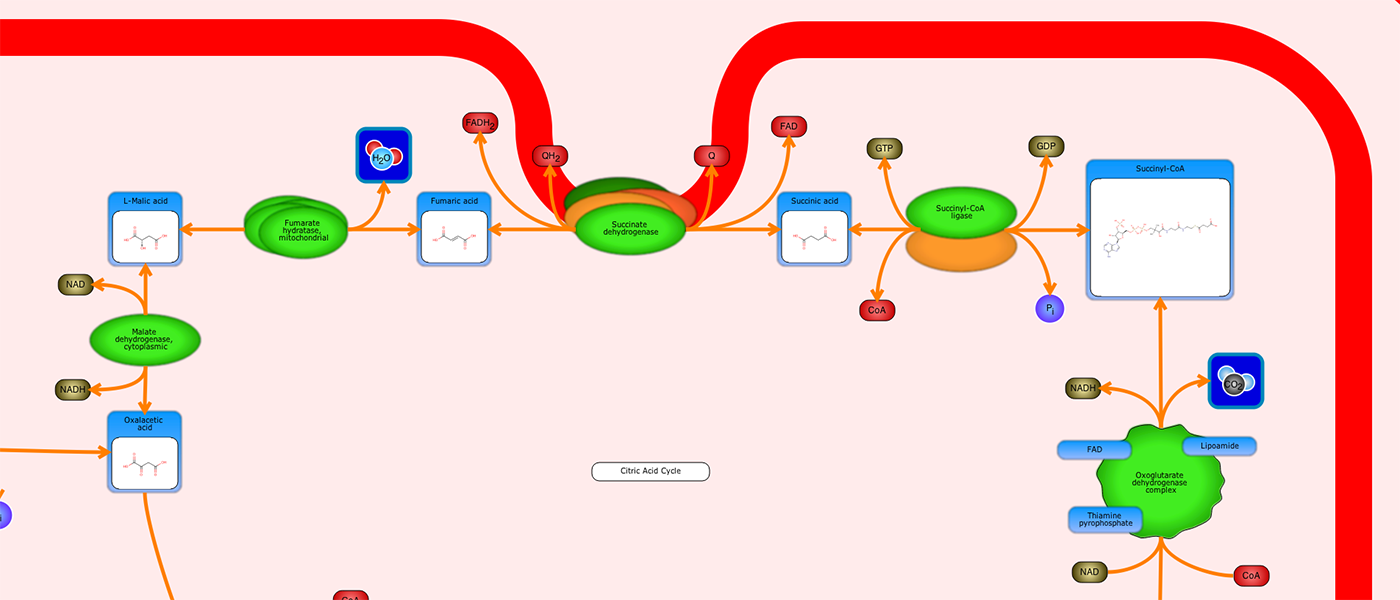
PathWhiz is a web server designed for the creation of colourful, visually pleasing, and biologically accurate pathway diagrams that are machine-readable and interactive. As a web server, PathWhiz is accessible from almost any place and compatible with essentially any operating system. It also houses a public library of pathways and pathway components that can be viewed and expanded upon by its users. PathWhiz supports a high level of biological detail and complexity, using a specially designed drawing palette to render metabolites (including automated structure generation), protein_complexes (including quaternary structures, covalent modifications, and cofactors), nucleic acids, membranes, subcellular structures, cells, tissues, and organs. Both small molecule and protein_complex/gene pathways can be constructed quickly and intuitively by combining processes such as reactions, interactions, binding events, and transports. Pathway replication functionality allows existing pathways to be used as a basis for the creation of new pathways and for existing pathways to be automatically propagated across species. PathWhiz supports the generation of BioPAX, SBGN-ML, SBML, and PWML exchange formats, in addition to high resolution PNG, image map, and SVG images that can be explored using its interactive viewer. PathWhiz has been used to generate over 700 pathway diagrams for a number of popular databases including HMDB, DrugBank and PathBank.
If you like this site, please follow us on Twitter and Facebook.
PathWhiz is supported by David Wishart, Departments of Computing Science & Biological Sciences, University of Alberta. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (Pathwhiz) and the original publication (see below). We ask that users who use this software cite the Pathwhiz paper in any resulting publications.
- Pon A, Jewison T, Su Y, et al. Pathways with PathWhiz. Nucleic Acids Res. 2015 Jul;43(W1):W552-9.
- Ramirez-Goana M, Marcu A, Pon A, et al. A Web Tool for Generating High Quality Machine-readable Biological Pathways. J Vis Exp. 2017 Feb 8;(120). Video here.
The developers of PathWhiz respect user privacy. Private pathway data will not be altered by anyone, including the PathWhiz web server administrators, without user permission, for as long as the web server is operational and as long as the user indicates the pathway data should remain private.